Could it really be true that previously unknown and undiscovered works of Shakespeare might be HIDDEN in vintage copies of Boys’ Life Magazine?
Let’s use METAGENOMICS find out!
Project Inspiration
I got the idea from a recent Youtube video by the creator Destin of “Smarter Every Day”, who showed how Metagenomics could be used to find the (King James) Apocrypha in old Ellery Queen mags.
Method Details
While there are many tools that are used for Metagenomics, Bioinformatics, Next Gen Genomic Assembly, aka, Metatranscriptonomics, for this project, we will be using the Open Source TRINITY suite:
https://github.com/trinityrnaseq/trinityrnaseq/wiki
When viruses are sequenced, using Next Gen Sequencing, raw samples are first contaminated with foreign DNA & RNA. Because, … well, why not! Makes it more interesting!
Then, the DNA or RNA is then “read” using Amplicon PCR using primers targeting the desired region of interest, and fed into Illumina©.
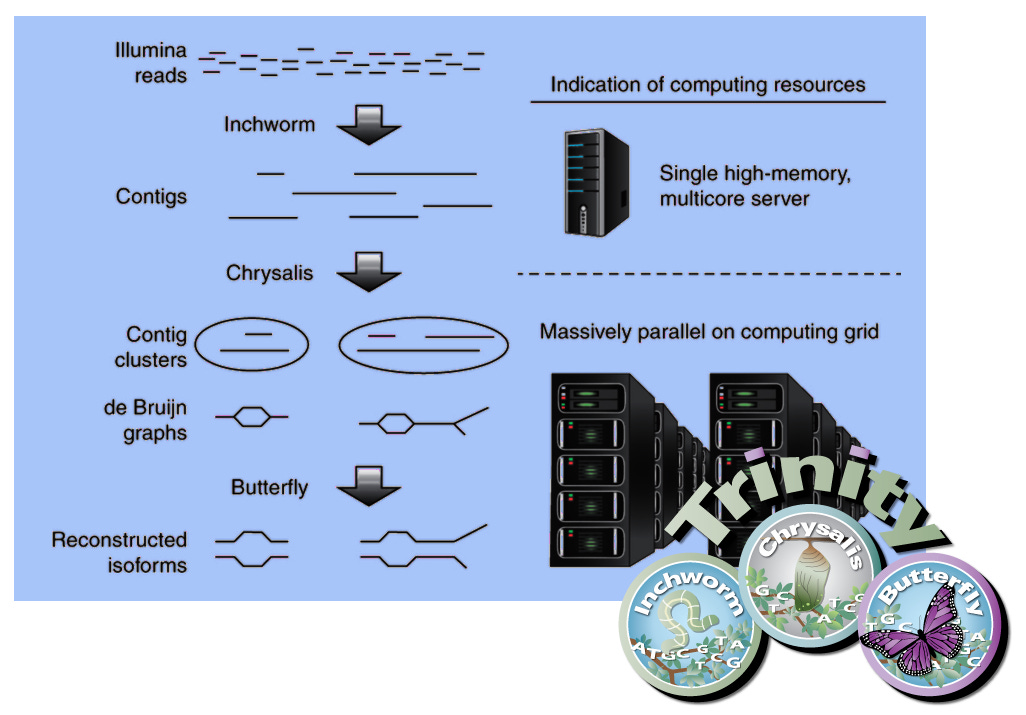
Inchworm® is used to transform the individual “reads” into contigs, based on statistical patterns found in existing viral genome databases.
Chrysalis℠ is then used to form Contig Clusters, and then formed into de Bruijn graphs.
Finally, Butterfly™ is used to produce the final reconstructed isoforms.
Here is an overview of the process:
The Shakespeare Modification
So instead of Bronchoalveolar lavage fluid (BALF) as the input to the process, we will be using a stack of vintage Boys’ Life magazines I found in my attic.
This will be a Mock Viral Cell Culture. Since biological contamination cannot be accomplished in a similar way, we have performed the following substitutions:
▶️ African Green Monkey Kidney Epithelial Cells (Vero):
Substitution: 👉 Popular Mechanics Magazine (1957 to 1969)
▶️ A549 (adenocarcinomic human alveolar basal epithelial) cells:
Substitution: 👉 Movie Classic Magazine (1954-1959)
▶️ Fetal Bovine Serum:
Substitution: 👉 Life Magazine (1962-1971, minus the Kennedy Assassination issue, missing from the source dataset)
▶️ Viral Growth Medium:
Substitution: 👉 A whole bunch of other random magazines that I found in the box.
How Our Method Differs
Instead of using pattern matching against existing viral genomes in GenBank, I have modified the source code to scan the existing works of Shakespeare collection at Project Gutenberg. (As a control, we seeded our AI with works of many other popular writers as well as Shakespeare)
The source magazines are clipped using a high-speed book scanner into short “Reads” of just a few words each.
These are fed into the Metagenomic Pipeline exactly as with Viral Sequencing. Then Contigs are formed according to statistical patterns found in the source texts.
We are doing the analysis on a Beowulf cluster of 256 Raspberry Pi Model 4’s running Ubuntu Linux/OpenHPC in a modified Torus Manifold / Hypercube configuration.
Here is a very promising Contig Cluster which found a (near perfect) match of Hamlet:

A Match!
(almost)
Building a Phylogenetic Tree
We ran the simulation 15M more times. In addition to finding 82% of Shakespeare’s catalogue, as well as 77 previously unknown sonnets, 3 plays, and 1 poem, we also found a large number of works by Ernest Hemmingway, PJ O’Rourke, e e cummings, Hunter S. Thompson, and Erica Jong, of all things.
This is Philogenetic Tree of our findings:
We will be posting detailed build notes first for Paid Subscribers.
SIGN UP NOW!
More soon…






I love the analogy. It's a riot! :)
How are you making it, Bill? Been kinda worried about ya...
Well, managing complex systems is your specialty. I know all this over exposure to in silico viruses would finally be contagious